An international collaboration has led to a breakthrough in inhibiting the function of the SARS-CoV-2 main protease, one of the key proteins of Covid-19. Although research began barely half a year ago, by applying the molecular LEGO concept, the consortium was able to identify more than 70 molecules that may serve as a good starting point for the development of drugs.
The Medicinal Chemistry Research Group of the ELKH Research Centre for Natural Sciences (TTK) is also participating in the research program, working in collaboration with research teams from the University of Oxford, the Diamond particle accelerator in England and the Weizmann Institute in Israel with the aim of finding new ways to treat COVID-19 infection by inhibiting the coronavirus proteins of SARS-Cov-2. The findings of the consortium have been published in the journal Nature Communication (https://www.nature.com/articles/s41467-020-18709-w) and the results have also highlighted in an editorial article (https://www.nature.com/articles/s41467-020-18710-3).
The research began with the isolation and purification of viral proteins, which resulted in the identification of a protein that is essential for viral replication, usually referred to as the SARS-Cov-2 main protease. This enzyme is responsible for the production of proteins that are important for the viability of the virus and which are encoded by the virus's genetic stock. This means that its effective inhibition prevents the virus from multiplying. In terms of treatment options, it is encouraging that the function of the virus's main protease is fundamentally different from that of human proteases. This means that the inhibitors developed are not expected to have the dangerous side effects of protease inhibition.
The research was based on the structure-based drug design already used successfully in the development of anti-viral drugs for the treatment of AIDS, where the three-dimensional structure of the target protein had to be determined first. Although the first protein structure was unveiled by German researchers in Science magazine on 24 April (DOI: 10.1126/science.abb3405), an international consortium that included Hungarian researchers was already working hard to identify promising new molecules. With this aim in mind, researchers have developed a new, efficient method based on the molecular LEGO concept, which enables efficient recognition of simple molecular building blocks, or fragments, that bind to a protein.
An essential element of the solution is that the fragments designed by the Hungarian research group not only find suitable cavities in the protein, but also react with the protein there, causing them to attach to the pockets of the protein. These molecules form a strong and lasting interaction that prevents the drug molecule from leaving the binding site, ensuring that the inhibition becomes permanent. The molecular sets developed by Hungarian, English and Israeli researchers were studied in the Diamond particle accelerator in England. The crystals of the protein were individually soaked in a solution of 1,250 different fragments, and the location of the protein-bound fragment 74 in the protease binding pocket was determined by X-ray diffraction.
-
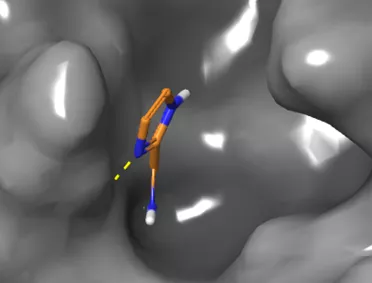
- A protein-bound fragment in the coronavirus major protease pocket
Thanks to the new procedure and particle accelerator, the measurements were completed in less than a month. Analysis showed that several of the tested Hungarian-developed reactive fragments proved to be effective. This can serve as a promising starting point and contribute to the development of new Covid treatments. One unique feature of the Hungarian building blocks is that the function of additional reference proteins was not inhibited, something which further reduces the risk of side effects. The Hungarian research group is already building on these promising fragments. This is supported not only by the Hungarian National Research, Development and Innovation Office but also by the UK Foreign Office.
The molecular LEGO concept
It has been more than a hundred years since Emil Fischer and Paul Erlich traced the effectiveness of drugs back to molecular interactions between molecules and proteins in the body. With the increase in knowledge concerning the structure of proteins, it was also found that drug molecules exert their effects by binding to the smaller or larger cavities of proteins. Identifying such cavities, as well as molecules capable of forming appropriate interactions within them, is one of the greatest challenges in the early stages of pharmaceutical research. Until the late 20th century, attempts were made to experimentally select molecules that fit into cavities from molecules previously produced for other purposes. Thanks to a new approach, the foundations of a molecular LEGO method based on testing and building molecules (fragments) that are significantly smaller than those of pharmaceuticals has been set out in the last ten years. With this method, the search for ligands begins with an examination of the binding of fragments and is based on the recognition that such molecules are more likely to bind to the cavities of proteins than larger molecules of drug candidate size. As a result, a starting point can be provided by screening a directory that already contains a few hundred or few thousand fragments. These starting points can be used for further development, taking into account the characteristics of the cavity, or in the event of several matches, may also lead to new candidates for medication.