In an international collaboration project led by Newcastle University, researchers explored the survival strategies of cold-tolerant bacteria and discovered a new so-called hibernation factor. Bálint Csörgő, the group leader of the MTA Momentum Gene Technology Lab at the Institute of Biochemistry, HUN-REN Biological Research Centre in Szeged (HUN-REN BRC Szeged), developed the necessary genome editing methodologies. The significant findings of the research were recently published in the leading scientific journal, Nature.

The overwhelming majority of our biological knowledge stems from the study of organisms found in our immediate surroundings or from within our own bodies. This holds true for bacteria as well, as the most well-studied species are from environments inhabited by humans or from within the human body itself. However, there are numerous conditions where the majority of these species are unable to survive but other bacteria are able to survive and even thrive. One such example is species belonging to the genus Psychrobacter, which can be found living in the icy terrain of Antarctica.
The primary goal of this research was to understand what kinds of special molecular mechanisms allow these cold-bearing bacteria to survive under such harsh conditions, specifically focusing on the basic biological process of protein translation. This international collaborative project identified a novel hibernation factor named Balon, which allows for ribosomes (the key machinery responsible for protein translation) to be put on “pause” under stressful freezing conditions, then quickly reactivated when conditions become more favorable. Binding of Balon protects ribosomes from damage during freezing conditions and allows for rapid commencement of translation when conditions improve.
The team of scientists first employed cryogenic electron microscopy to study the ribosomes of these hibernating bacteria, a technique capable of determining the structure of biomolecules at high resolution.
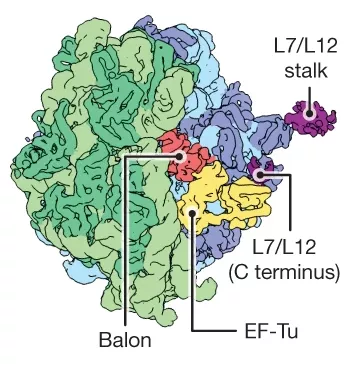
These investigations first showed the presence of Balon, which was later characterized in greater detail using other complementary methods, including mass spectrometry. To identify and characterise the gene encoding Balon, genome editing methodologies needed to be developed that could be used to genomically modify different Psychrobacter species, which are otherwise difficult to cultivate under laboratory conditions.
This task was carried out by Bálint Csörgő, group leader at HUN-REN BRC Szeged. Using the techniques developed in his lab, targeted gene knockouts of Balon were generated in multiple Psychrobacter species and these strains displayed an impaired capacity to withstand cold stress. When searching for similar genes in other bacterial organisms in genomic databases, many different species were identified carrying homologues of the gene, indicating a potential role for Balon in general stress responses. Interestingly, Balon also showed similarity to a general translation factor of higher-order eukaryotes, indicating an important evolutionary role for the protein.
This research highlights the importance of going beyond common model organisms and studying less well-known extremophiles, as novel aspects of basic cellular processes can still be discovered.