László Nagy, senior research fellow at the Synthetic and Systems Biology Unit at the Biochemistry Institute of the HUN-REN Biological Research Centre, Szeged (HUN-REN BRC) led the international collaboration targeted to mapping up the consequences of contaminated genomic data. The Hungarian researchers developed a sequence cleaning software that is significantly more accurate in filtering out and removing contaminants from genomic data than previous methods. The paper presenting the results was published in the prestigious Nature Communications journal.
The joint study by researchers of the HUN-REN BRC, Joint Genome Institute (JGI, USA) and the National Energy Research Scientific Computing Center (NERSC, USA) explores the potential consequences of genomic data contaminated with foreign sequences that have been erroneously uploaded to sequence databases, and the cleanup of these contaminants.
The researchers from Szeged have developed an entirely new computer software (ContScout), which identifies and removes contaminants lurking in genomic data (Figure 1) with significantly greater accuracy compared to solutions previously published by other groups, while leaving genes acquired through horizontal gene transfer untouched.
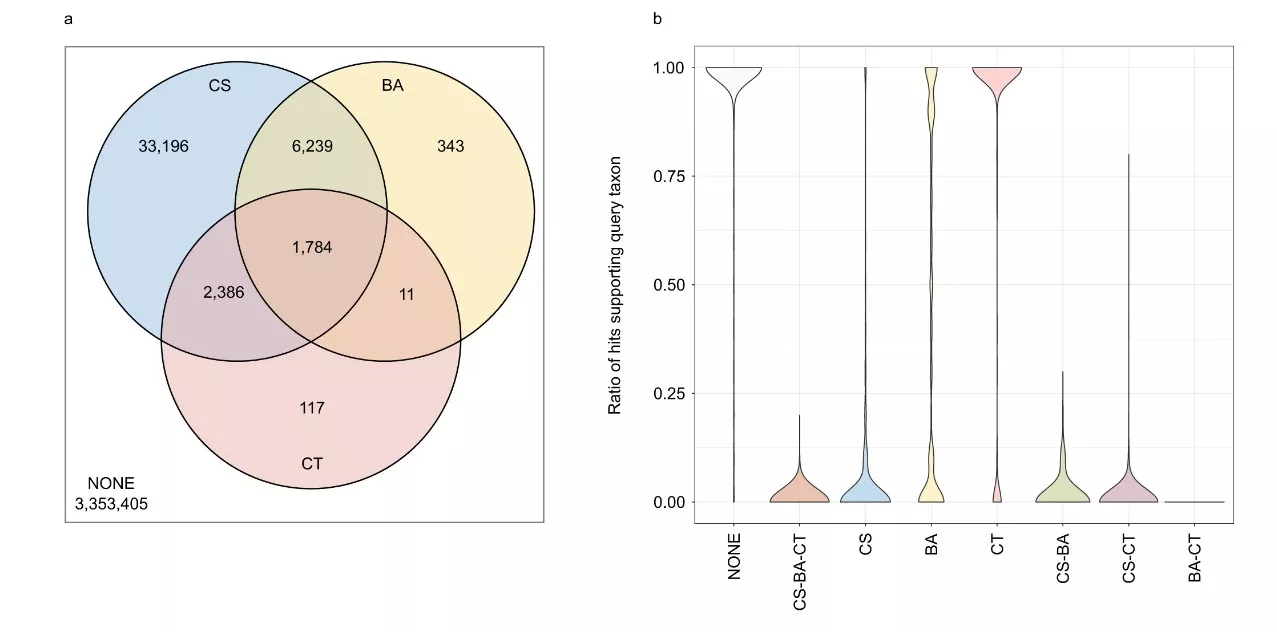
Figure 1. Comparison of the ContScout program with software previously published by other research groups. The Venn diagram (a) and the violin plot (b) contain data for the 200 most contaminated genomes. Abbreviations: NONE – No Contamination. CS: Contaminant protein identified by ContScout, BA: Contaminant protein identified by BASTA, CT: Contaminant protein identified by Conterminator.
The researchers detected some degree of contamination in more than half of the 844 examined eukaryotic genomes, with most of the contaminants originating from bacteria. In some cases, the contamination was so extensive that the published genomic data allowed for the reconstruction of a nearly complete genome of a second organism.
The paper demonstrates through detailed examples the distorting effect that contaminant sequences have on experiments aimed at uncovering the unique gene families and the evolutionary history of organisms.
The researchers involved in the collaboration hope that their results, published in a prestigious journal, will successfully draw the scientific community's attention to the importance of removing genomic contaminants and that the developed sequence cleaning software can become part of the generally accepted genomic analysis workflows.