Roland Wirth and his colleagues at the Institute of Plant Biology of the ELKH Biological Research Centre, in the Microbial and Algal Genomics Research Group (including the Extracellular Electron Transfer Momentum Group) led by Gergely Maróti, in close collaboration with the Biotechnology Department of the Faculty of Science and Informatics of the University of Szeged have investigated the interactions of key microorganisms involved in biogas production - methanogenic archaea, hydrolytic and syntrophic bacteria - as well as the stability and functions of microbial communities in industrial-scale anaerobic degradation systems, real biogas plants. In their study the researchers demonstrated that although methanogenic archaea have low relative abundance in the system, they are by far the most active microorganisms in biogas fermenters. Methanogens, which utilize hydrogen and carbon dioxide, are dominant in the community and closely interact with representatives of hydrolytic Firmicutes and Bacteroidota bacterial strains. The study highlights that the diversity of the methanogenic population is crucial for efficient biogas production. The paper describing the results of the research employing state-of-the-art metagenomic and metatranscriptomic approaches, were published by the authors in The ISME Journal.
Biogas production is based on the efficiency of organic matter degradation. In this process, a complex microbial community participates, with each microorganism having a specific role. The composition and activity of the microbial community are influenced by various factors, such as microbial interactions, biomass composition, and metabolites generated during anaerobic degradation. Unraveling microbial interactions and their mechanisms is a complex task, as most microorganisms cannot be studied in the laboratory without their network of partners. Therefore, studying syntrophic interactions in complex communities requires a specialized approach. Advanced sequencing technologies and bioinformatic algorithms can offer solutions to this problem. They enable the reconstruction of complete genomes of individual microbial species and the identification and quantitative characterization of functions associated with specific microorganisms, even in complex communities such as biogas-producing systems. In this research the authors utilized a genome-centric approach employing artificial neural networks for the deep analysis of metagenomic data from industrial-scale biogas reactors, allowing for the reconstruction of numerous bacterial and archaeal genomes.
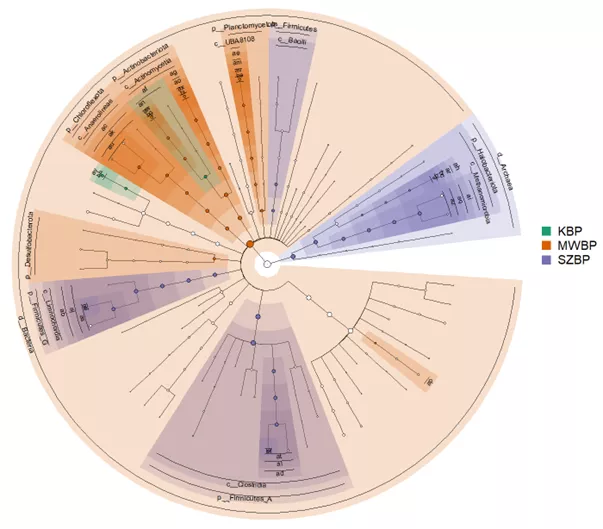
Figure 1. Characteristic microorganisms identified in the three biogas plants
The results of the study have shed light on our incomplete understanding of the activities of biogas-producing communities and the relationships among the microorganisms residing in these communities. The researchers identified numerous previously unknown microbes and also explored the activity, functional roles and interactions of these microorganisms with others. They identified metabolic pathways crucial for biomass-to-methane conversion between methanogenic archaea and syntrophic bacteria partners. The study's findings also highlighted the significant role of methanogenic microbial diversity in maintaining the functional stability of the community.
Publication:
Wirth R., Bagi Z., Shetty P., Szuhaj M., Cheung TTS., Kovács KL. and Maróti G. (2023). Inter-kingdom interactions and stability of methanogens revealed by machine-learning guided multi-omics analysis of industrial-scale biogas plants. The ISME Journal. DOI: 10.1038/s41396-023-01448-3